Image data is hosted using the open-source NeuroInformatics Database (NiDB). NiDB is designed to store, retrieve, analyze, and share neuroimaging data. Modalities include MR, EEG, ET, video, genetics, assessment data, and any binary data. Subject demographics, family relationships, and data imported from RedCap can be stored and queried in the database.
- Store any research imaging data, including MR, CT, EEG, ET, Video, Task, GSR, Consent, MEG, TMS, and more
- Store any assessment data (paper-based tasks)
- Store clinical trial information – manage data across multiple days & dose times
- Built-in DICOM receiver. Send DICOM data from PACS or MRI directly to NiDB
- Bulk import of imaging data
- User and project based permissions, with project admin roles
- Search and manipulate data from subjects across projects
- Automated imaging analysis pipeline system
- “Mini-pipeline” module to process behavioral data files and extract timings
- All stored data is searchable. Combine results from pipelines, QC output, behavioral data, and more in one query
- Export to NFS, FTP, Web download, NDA (NIMH Data Archive format), or export to a remote NiDB server
- Export data in squirrel, BIDS, Nifti, and anonymized DICOM formats
- Project level checklists for imaging data
- Automated motion correction and other QC for MRI data
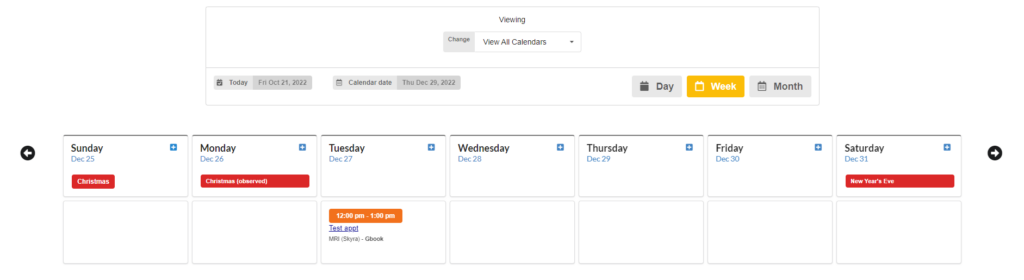
- Calendar for scheduling equipment and rooms
- Usage reports, audits, tape backup module
- Intuitive, modern UI. Easy to use
- .rpm based installation for CentOS 8, RHEL 8, Rocky Linux 8 (not for CentOS Stream)
Featured Features
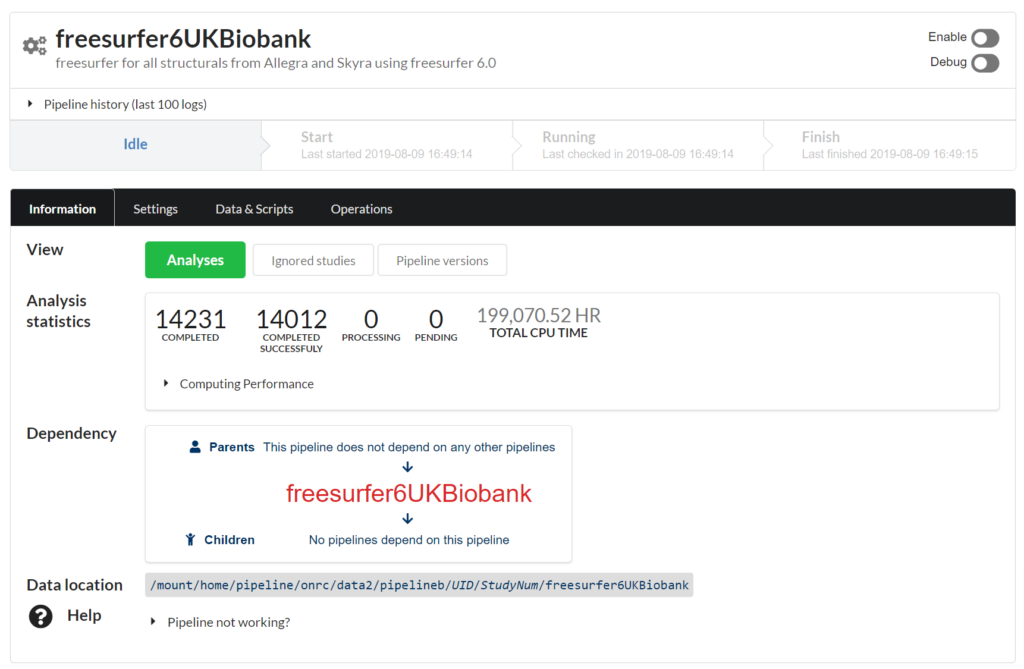
Full analysis pipeline system
From raw data to analyzed, and storing result values/images. Utilize a compute cluster to process jobs in parallel.
Example analysis : 200,000 hrs of compute time completed in a few weeks. Hundreds of thousands of result values automatically stored in NiDB and are searchable.
Store any type of data
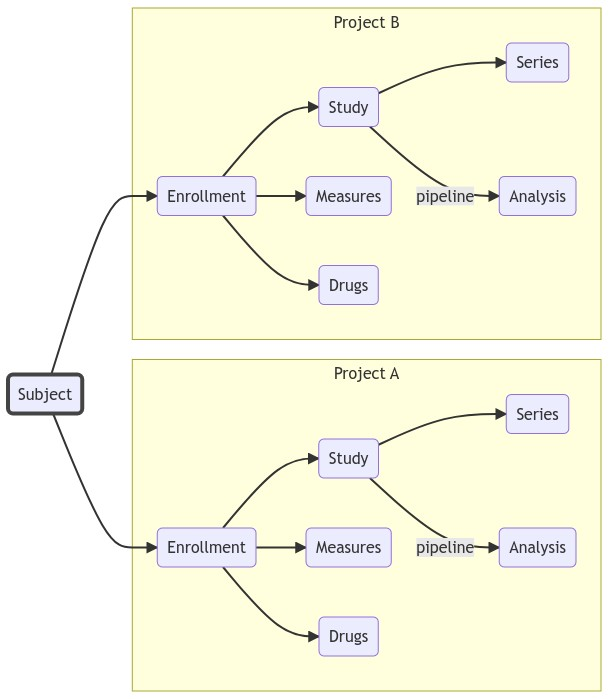
Any type of imaging data: binary; assessment; paper based; genetics. See full list of supported modalities. All data is stored in a hierarchy: Subject –> Study –> Series. Data is searchable across project and across subject.
Automated import of DICOM data
DICOM data can be automatically imported using the included dcmrcv receiver. Setup your MRI or other DICOM compatible device to send images to NiDB, and NiDB will automatically archive them. Image series can arrive on NiDB in any order: partial series, or full series to overlap incomplete series.

Clinical trials
NiDB stores multiple time-points with identifiers for clinical trials; exact day numbers (days 1, 15, 30 …) or ordinal timepoints (timepoint 1, 2, 3 …) or both (day1-time1, day1-time2, day2-time1, … )
Bulk import of imaging data
Got a batch of DICOMs from a collaborator, or from an old DVD? Import them easily.
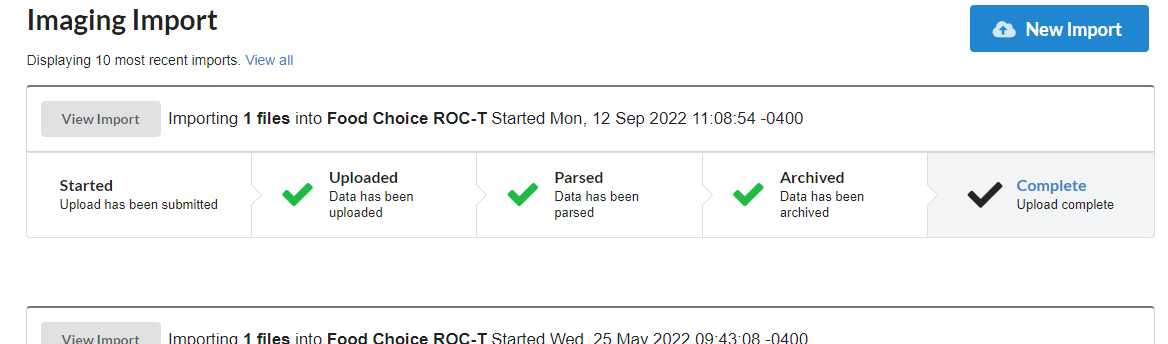
Search and export imaging data
Find imaging data across projects and export data. Search by dozens of criteria.
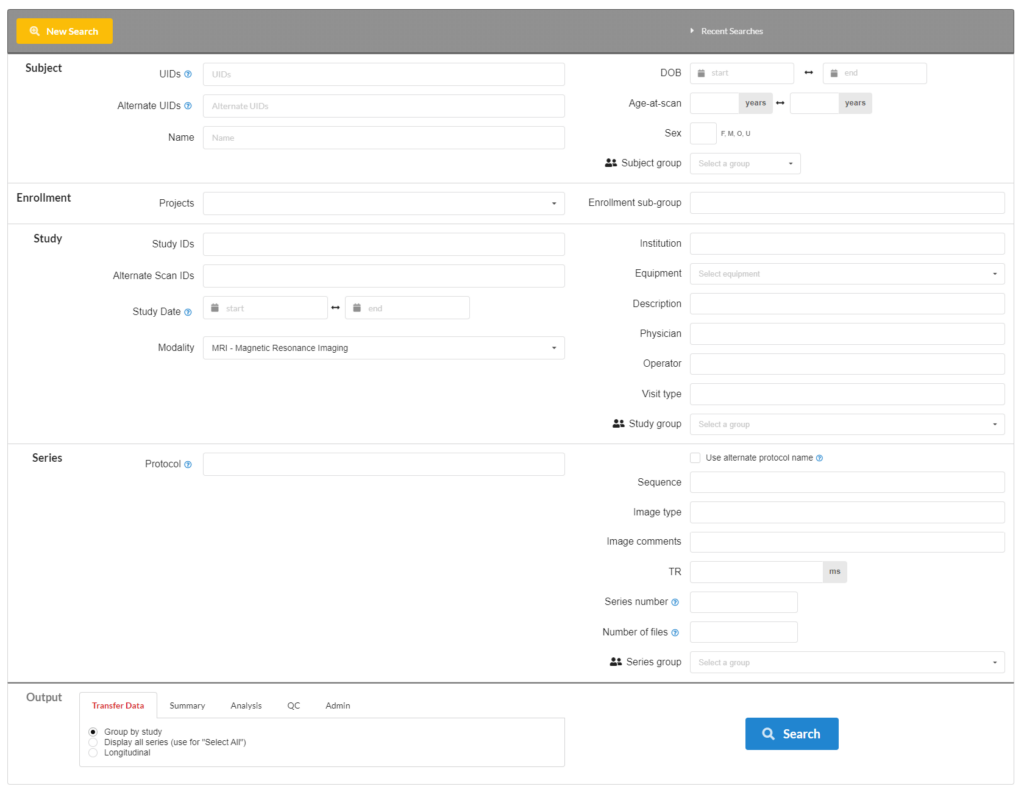
Export Destinations
- Linux NFS share
- Web
- Public download/dataset
- Local FTP
- Remote NiDB instance
Package formats
- squirrel
- BIDS
- NDA
Export image formats
- Original raw data (DICOM, Nifti, Par/Rec)
- Anonymized DICOM
- Nifti 3D (.nii & .nii.gz)
- Nifti 4D
Search and export non-imaging data
Data obtained from pipeline analysis, imported from redcap, computed measures, drugs, vitals, measures, are all searchable.
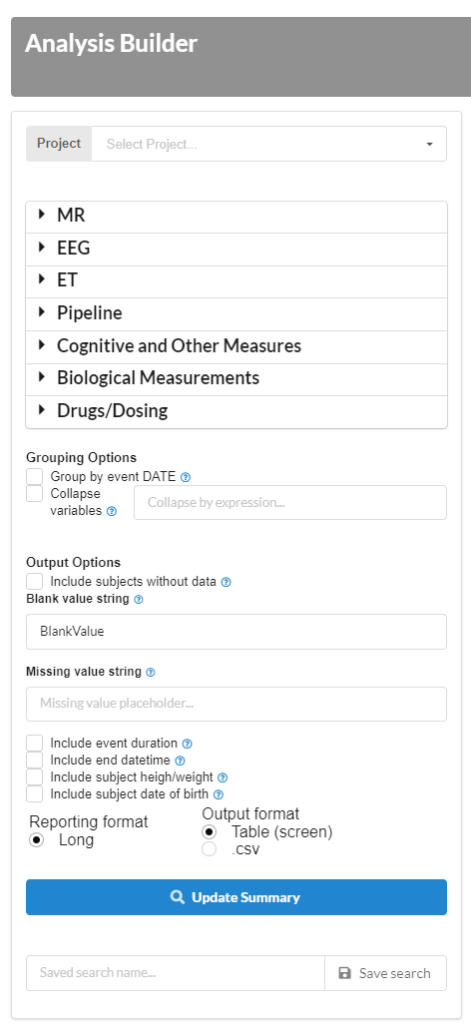
Automated MR quality control
Automatically generated metrics, exportable as .csv and tables.
Calendar
Manage your resources with a fully featured calendar, running securely on your internal network. Repeating appts, blocking appts, and time requests.